Turn on suggestions
Auto-suggest helps you quickly narrow down your search results by suggesting possible matches as you type.
Showing results for
Data Engineering
Turn on suggestions
Auto-suggest helps you quickly narrow down your search results by suggesting possible matches as you type.
Showing results for
- Databricks
- Data Engineering
- [SOLVED] maxPartitionBytes ignored?
Options
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Options
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
10-22-2021 06:17 AM
Hello all!
I'm running a simple read noop query where I read a specific partition of a delta table that looks like this:
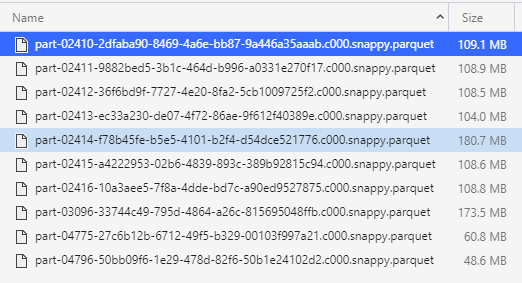
When I configure "spark.sql.files.maxPartitionBytes" (or "spark.files.maxPartitionBytes") to 64MB, I do read with 20 partitions as expected. THOUGH the extra partitions are empty (or some kilobytes)
I have tested with "spark.sql.adaptive.enabled" set to true and false without any change in the behaviour.
Any thoughts why this is happening and how force spark to read in smaller partitions?
Thank you in advance for your help!
Labels:
- Labels:
-
Delta table
1 ACCEPTED SOLUTION
Accepted Solutions
Options
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
10-23-2021 02:36 AM
The AQE will only kick in when you are actually doin transformations (shuffle/broadcast) and it will try to optimize the partition size:
AFAIK the read-partitionsize is indeed defined by maxPartitionBytes.
Now, I do recall a topic on stackoverflow where someone asks a similar question.
And there they mention the compression coded also matters.
Chances are you use snappy compression. If that is the case, the partition size might be defined by the row group size of the parquet files.
https://stackoverflow.com/questions/32382352/is-snappy-splittable-or-not-splittable
http://boristyukin.com/is-snappy-compressed-parquet-file-splittable/
Also David Vrba mentions the compression used too:
7 REPLIES 7
Options
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
10-22-2021 07:35 AM
How did you determine the number of partitions read and the size of these partitions?
The reason I ask is because if your first read the data and then immediately wrote it to another delta table, there is also auto optimize on delta lake, which tries to write 128MB files.
(spark.databricks.delta.autoCompact.maxFileSize)
Options
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
10-22-2021 02:48 PM
Hello Werners,
I'm looking at the input size of each partion at the stage page of the spark UI. As I said I did a noop operation, there is no actual writing. My goal it to have control on the partition size at read which the conf I'm playing with is supposed to do
Options
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
10-22-2021 12:56 PM
AQE doesn't affect the read time partitioning but at the shuffle time. It would be better to run optimize on the delta lake which will compact the files to approx 1 GB each, it would provide better read time performance.
Options
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
10-22-2021 02:51 PM
Hello Ashish,
I was just wondering as AQE might change the expected behaviour. As stated before, my issue here is to control the partition size at read not to optimise my reading time.
Why it correctly breaks the 180MB file in 2 when 128 is the limit, but not the 108 MB files when the limit is 64
Options
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
10-23-2021 02:36 AM
The AQE will only kick in when you are actually doin transformations (shuffle/broadcast) and it will try to optimize the partition size:
AFAIK the read-partitionsize is indeed defined by maxPartitionBytes.
Now, I do recall a topic on stackoverflow where someone asks a similar question.
And there they mention the compression coded also matters.
Chances are you use snappy compression. If that is the case, the partition size might be defined by the row group size of the parquet files.
https://stackoverflow.com/questions/32382352/is-snappy-splittable-or-not-splittable
http://boristyukin.com/is-snappy-compressed-parquet-file-splittable/
Also David Vrba mentions the compression used too:
Options
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
10-25-2021 08:42 AM
Thanks
You have a point there regarding parquet. Just checked, it does read a separate row group in each task, the thing is that the row groups are unbalanced, so the second taks gets just some KB of data.
Case closed, kudos @Werner Stinckens
Options
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
10-26-2021 01:45 PM
Hi @Pantelis Maroudis
Thanks for your reply. If you think @Werner Stinckens reply helped you solve this issue, then please mark it as best answer to move it to the top of the thread.
Announcements
Welcome to Databricks Community: Lets learn, network and celebrate together
Join our fast-growing data practitioner and expert community of 80K+ members, ready to discover, help and collaborate together while making meaningful connections.
Click here to register and join today!
Engage in exciting technical discussions, join a group with your peers and meet our Featured Members.
Related Content
- value is null after loading a saved df when using specific type in schema in Data Engineering
- Ingesting complex/unstructured data in Data Engineering
- Dashboard update through API in Administration & Architecture
- Dashboard update through API in Data Engineering
- synapse pyspark delta lake merge scd type2 without primary key in Data Engineering